How to calculate melting temperature of DNA "2+4 rule of thumb"
Вставка
- Опубліковано 14 лип 2024
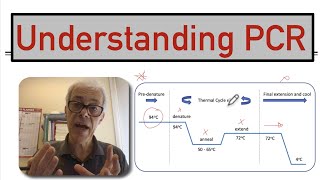
- The melting temperature (Tm) is the temperature at which one-half of a particular DNA duplex will dissociate and become single strand DNA. The stability of a primer-template DNA duplex can be measured by its Tm. Primers with melting temperatures in the range of 52-58°C generally produce better results than primers with lower melting temperatures. While the annealing temperature can go as high as 72°C, primers with melting temperatures above 65°C have a higher potential for secondary annealing.
Primer length and sequence are of critical importance in designing the parameters of a successful amplification. The melting temperature of a nucleic acid duplex increases both with its length, and with increasing GC content. A simple formula for calculation of the (Tm) is:
Tm = 4(G + C) + 2(A + T) °C
The actual (Tm) is influenced by the concentration of Mg2+, K+, and cosolvents. There are numerous computer programs to assist in primer design. The formula given above for (Tm) is simplistic; there are many primer design programs which use more complex nearest-neighbor thermodynamics values








extremely helpful!! THANK YOU.
That was helpful! :)
thank you very much it's clear enough and helped my in my exam
Informative
Super Helpful!
Glad you think so!
Nice..🎉
Thanks 😊
If you are faced with a question that shows the sense strand and the anti sense strand and asks for the melt, should you count all the nucleotides on both strands to find the melt, or just one?
No need
1. If we know the sequence of one strand - we instantly know the sequence of the other.
2. Every base have only 2 or 3 hydrogen bonds if we know th sequence of one strand we can calculate number of hydrogen bonds - other strand is not needed.
Watch my new short - INDIANS with BLUE eyes? Sereously? ua-cam.com/users/shortsKhdARdHoI9o
I was about to sleep due to his voice
This is one of my first videos. Lol
why the 2 and 4 in the formula?
I'm questioning the same
The "2" and "4" portions of the equation are weighted factors.
More weight are given to the GC term of the equation than the AT term because GC forms 3 hydrogen bond while AT forms only 2 Hydrogen Bonds.
Watch my new video " How to develop SUPER memory" ua-cam.com/video/9k2sM_jcZUQ/v-deo.html
Percentage formula
No one is doing this manually - everything is done by sophisticated software.. This video just provides insights on basic principles of DNA melting point calculation.
And the pourcentage?
Watch my new video " How to develop SUPER memory" ua-cam.com/video/9k2sM_jcZUQ/v-deo.html
Thanks for trying to help and teach to the large community. But this method is very very bad. It just does not work. Never. Melting temperature depends strongly on the concentration of nucleic acid and salt. Not just Mg2+, but even more of sodium. It even depends on the order of the nucleotides. The same number of A,T,G,C in different orders have different melting temperatures.
And by the way, the melting temperature does NOT depend on the hydrogen bonds between strands (not only anyway). Some combinations of mismatches even increase the temperature! (for instance a TT mismatch is sometimes stronger than the matching AT).
The so-called Wallace method is one of the hardest myth to debunk in molecular biology. Please people DO NOT USE 4(G+C)+2(A+T). THIS DOES NOT WORK. EVER. There are plenty of super easy to use software around, based on the correct way of doing things, called "nearest-neighbors". I am not going to quote them (mostly because I am the author of one ;-) )
Thank you for your comment, but this video is for the entry level course, and Idea was just to familiarize audience with this method of calculation. Software calculations are too complicated to explain in 10 minutes video.
I understand that Nikolay. But the method is wrong. It does not work. It does not predict anything close to the melting temperature. So it is exactly like a primary school teacher teaching kids that 2+2=5 because it is a "close approximation". I know you meant well, and I know that you learnt about this method in some textbooks that have propagated this myth for a few decades without checking. But it is just that. A myth. The method did work for some molecular biologist 60 years ago, who worked in some very specific conditions, always the same. And for some reason, mostly because it looks simple and easy to remember, it has been propagated as a meme. This is total nonsense, and "teaching" that does not help at all. Really. Just try it yourself on actual data (actual measured melting temperatures. There are plenty in the literature. I can send you papers if you need). I am sorry to pick on you, because I really appreciate people who teach science through UA-cam and other social media. But this is not science. Sorry.
Scientist have to start with something - when structure of DNA was discovered - this gave Idea about forces that hold two strands together - and it was obvious hydrogen bounds. So next step was just to calculate how many hydrogen bounds we have in specific DNA fragment and try to calculate melting temperatures. Next step in experiments was to calculate melting points of say, fragments consisting of bases with 3 bounds, 2 bounds and mixtures 2-3 bounds. Then with different mixtures say A-T-A-T and AA-TT All these experiments lead to new correlation formulas that took into account not just a number of bases and number of bounds but a Sequence of the bases along with other factors such as chemical concentrations and so on. But we have to mention that all these later "adjustments" to the initial formula changed it beyond recognition and nowadays so complicated that nobody do such calculations "on the knees", especially when we have a mixture of different primers - the more primers we have the more complicated calculations become especially in such cases as forensic DNA kits when we have mixture of at least 28 different primers - that have to be unique to 28 different sequence of DNA and should at the same time have the same melting and annealing temperatures. It is impossible to imagine to do such calculations manually.
Such chemical produces - gigants as Sigma Aldaich give a credit to Wallace www.sigmaaldrich.com/technical-documents/articles/biology/oligos-melting-temp.html
And this company know their business.
But you right - I should probably have to mention that this video is just historical review and not something that is used in modern daily lab routine.
Most of us are from India Right???
No.
@@GeneticsLessons most of the viewers in comments section are from India
Yes.