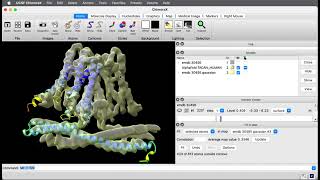
How to predict a large protein structure with AlphaFold using ChimeraX
Вставка
- Опубліковано 16 тра 2023
- To predict a large protein complex (more than 1200 amino acids) with AlphaFold using ChimeraX you need to use an A100 graphics processor (GPU) on Google Colab. This video shows you how to choose the A100 GPU with 40 GB of memory. You will only be able to use the A100 GPU if you pay Google for "compute units" on Google Colab.
- Наука та технологія








Is there a difference in GPU resource access when using pay-as-you-go versus subscription? I wonder because after buying 100 units without a subscription, the A100 was unavailable (although I could select it). Thanks.
I tried using colab for multimer . I paid for colab pro . Structures, plot everything were done but it's not downloading. ! In one structure (1700 amino acids) , I lost 50 units out of 100. And there is no visible solution to get the zip file. . If you know any solution for it, would u please share .
Does anyone know if you can use ChimeraX with Autodock Vina
🎉🎉🎉